New version of May 27, 2012:
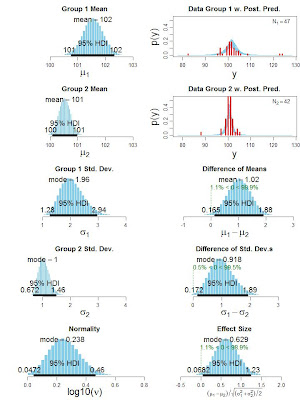
Bayesian estimation for two groups provides complete distributions of credible values for the effect size, group means and their difference, standard deviations and their difference, and the normality of the data. The method handles outliers. The decision rule can accept the null value (unlike traditional t tests) when certainty in the estimate is high (unlike Bayesian model comparison using Bayes factors). The method also yields precise estimates of statistical power for various research goals. The software and programs are free, and run on Macintosh, Linux, and Windows platforms. See this linked page for the latest paper and the software.
Search tag: BEST (for Bayesian estimation)
(This is the second revision of a the first version announced at a previous post.)
This comment has been removed by the author.
ReplyDeleteThanks for your excellent work in making this so accessible! At line 67 of BESTexample.R I get one plot but also this message:
ReplyDeleteError in file(filename, "r", encoding = encoding) :
cannot open the connection
In addition: Warning message:
In file(filename, "r", encoding = encoding) :
cannot open file 'plotPost.R': No such file or directory
I found plotPost.R over here http://www.indiana.edu/~kruschke/DoingBayesianDataAnalysis/Programs/plotPost.R and that made it all work fine.
It might be helpful to include a comment in the BESTexample.R about where to find this, or bundle it in with the zip file for completeness.
Yikes! Thank you for notifying me of this problem. I had simply zipped the wrong plotting file into the package. I have now deleted the wrong file and replaced it with the correct one, so I think that the zip file really is complete now. Thank you again!
ReplyDeleteI made an online version on the BEST routine. Maybe not to be used for serious analysis but quick n' easy!
ReplyDeleteCheck it out at:
http://www.sumsar.net/best_online/
Galeri B: Thanks again for the great web app. Here's the blog post:
ReplyDeletehttp://doingbayesiandataanalysis.blogspot.com/2013/01/bayesian-estimation-in-new-web-app.html
Hi, John-
ReplyDeleteI'm fairly new to Bayesian analyses and trying to work through your text. I'm having some trouble doing something similar to BEST, but for paired data, where individuals are observed in both groups being compared. I'm trying to visualize such a model using a hierarchical model diagram, but that gets a bit confusing to me. Any hints?
Much thanks!
Colin
Hi Colin.
ReplyDeleteA typical approach to paired data is to take the differences and treat the differences as a single group. Then do a single-group analysis on the difference scores. The BEST software now includes a one-group version; see
this blog post:
http://doingbayesiandataanalysis.blogspot.com/2012/09/one-group-version-of-best-bayesian.html
BEST wishes!
Dear John,
ReplyDeleteI am having a recurring error on plotting HDI in R studio on using BEST where it is saying compval is not a valid graphical parameter..
plot (BESTout, compvalue =1 , ROPE= c(-0.1,0.1))
Warning messages:
1: "compval" is not a graphical parameter
can you point out what is the source of this error and how can i correct it..
Regards,
Anupam K Singh
Department of Medicine
RIMS,Ranchi,India
Dear Anupam:
ReplyDeleteThere are two sets of programs for BEST in R. One set is the original that I created. A second set is an R package created by Mike Meredith that better implements the functionality in R-style commands, but the arguments and output are a bit different than in my original programs.
In my original programs, there are distinct functions for the two-group case and the one-group case. the arguments of the two-group plotting function are shown here:
> source("BEST.R")
> BESTplot
function( y1 , y2 , mcmcChain , ROPEm=NULL , ROPEsd=NULL ,
ROPEeff=NULL , showCurve=FALSE , pairsPlot=FALSE ) { ...
Notice that BESTplot (above) has no comparison-value arguments because the function assumes the comparison value should be zero.
The arguments of the one-group plotting function are shown here:
> source("BEST1G.R")
> BEST1Gplot
function( y , mcmcChain , compValm=0.0 , ROPEm=NULL ,
compValsd=NULL , ROPEsd=NULL ,
ROPEeff=NULL , showCurve=FALSE , pairsPlot=FALSE ) { ...
Notice that BEST1Gplot (above) has two different comparison-value arguments, one for the mean (namely, compValm) and one for the standard deviation (namely, compValsd).
In Mike Meredith's sophisticated re-packaging of the programs, the two-group and one-group cases are combined. You can plot the various parameters singly using the plot command, or plot the parameters together using the plotAll command. In RStudio, you can find out about the functions by looking in the sub-window that has a "Packages" tab. Scroll down the list of packages until you find BEST. Click it, and information will open in the Help tab, including a list of functions in the BEST package. Here are the arguments of the plotting functions:
> library("BEST")
> plotAll
function (BESTobj, credMass = 0.95, ROPEm = NULL, ROPEsd = NULL,
ROPEeff = NULL, compValm = 0, compValsd = NULL, compValeff = 0,
showCurve = FALSE, ...)
plot(x, which = c("mean", "sd", "effect", "nu"), credMass = 0.95,
ROPE = NULL, compVal = 0, showCurve = FALSE, ...)
Notice that the (method for the) plot function has an argument named compVal for specifying the comparison value. The plotAll function has arguments compValm and compValsd and compValeff.
P.S. Mike Meredith produced an extensive manual that explains examples of his BEST package far better than I did in the previous comment. Please take a look here:
ReplyDeletehttp://cran.r-project.org/web/packages/BEST/BEST.pdf
Dear Dr. Kruschke
ReplyDeleteThank you for your reply, I was able to reanalyse my data set with your help. It is much better to present data meaningfully with bayesian methods. I have been planning toreanalyse some of my old publications data with Bayesian methods .could you help me in resolving these queries?
1) a recent publication of mine evaluates comparative accuracy of gene-xpert(known prior sens, 75-90 % n specificity 95-100 %) and sputum microscopy( known prior sens, 40-60 % n specificity 95-100 %)...on the study of 500 pts with 120 true pos BACTEC as gold std...96/120 were gene expert positives...compared to 73/120 sputum positives..6 false positives in both groups..
I have already done ROC curves n other frequentist analysis. on the group...just wanted to know if there is a more meaningful Bayesian way to present my result..a helpful Reference or an R code would help..as I am an end user of R..and newbie...have just started studying Bayesian stats with no instruction .n my programming skills are pretty poor
how do we put priors to do a meaningful sensitivity analysis according to varying priors
2) how do we compare two rates eg. death rates n hospital admission percentage etc..when we compare two methods...by Bayesian methods..any R code for that..or a good reference would help
In one of my papers studying effect of metformin on B 12 deficiency...in diabetics
30/82 diabetics on metformin had b 12 deficierncy while 0nly 8/62 diabetics not on metformin had b 12 deficiency. previous known prevalence of b 12 def in metformin taking diabetics is between 20-30 percent while in diabeticsd not taking metformin is around 5 -10 percent...i have already done comparison of proportions n chi square..i want to know if i can use bayes to make it more meaningful. ?..how do we include knoweledge of priors in bayesian analysis..of comparisdon of proportion or chi square..
Anupam kumar Singh
MD, Chief Resident, Department Of Medicine
RIMS, Ranchi,India