Within days of each other I received two emails asking about Bayesian estimation of correlations and differences of correlations. Hence this post, which presents an implementation in JAGS and R.
Traditionally, a joint distribution of several continuous/metric variables is modeled as a multivariate normal. We estimate the mean vector and covariance matrix of the multivariate normal. When standardized, the covariance matrix is the correlation matrix, so pairwise correlations can be inspected. Moreover,
if it's meaningful to compare correlations, then we can also examine the posterior difference of pairwise correlations.
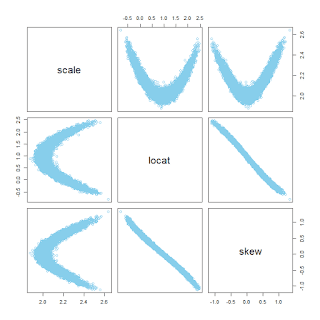
As an example, I'll use the data regarding Scholastic Aptitude Test (SAT) scores from Guber (1999), explained in Chapter 18 of
DBDA2E. We'll consider three data variables: SAT total score (SATT), spending per student (Spend), and percentage of students taking the exam (PrcntTake). Here's a plot of the data from different perspectives:
Instead of doing linear regression of SAT on Spend and PrcntTake, we'll describe the joint distribution as a multivariate normal. This is straightforward in JAGS, because JAGS has a multivariate normal distribution built in. The model specification includes the likelihood function:
zy[i,1:Nvar] ~ dmnorm( zMu[1:Nvar] , zInvCovMat[1:Nvar,1:Nvar] )
where the "z" prefixes of the variable names indicate that the data have been standardized inside JAGS. In the code above,
zy[i,:] is the
ith data point (a vector),
zMu[:] is the vector of estimated means, and
zInvCovMat[:,:] is the
inverse covariance matrix. Just as JAGS parameterizes the normal distribution by the
inverse variance (i.e., the precision), JAGS parameterizes the multivariate normal by the inverse covariance.
The prior on each mean is a broad normal. The prior on the inverse covariance matrices is a generic Wishart distribution, which is a multidimensional generalization of a gamma distribution. The Wishart is a distribution over matrices. The Wishart is built into JAGS.
The R code appended below provides the complete script, including the model specification and the graphics commands. The script needs the utilities script from DBDA2E for opening graphics windows.
Here is some selected output from the program. In particular, the program produces plots of the posterior distributions of the pairwise correlations, presented with the data and level contours from the multivariate normal:
Finally, here is a plot of the posterior of the difference of two correlations:
You should consider differences of correlations only if it's really meaningful to do so. Just as with taking differences of standardized regression coefficients, taking differences of correlations is assuming that the correlations are "on the same scale" which only makes sense to the extent that one standard deviation of SAT is "the same" as one standard deviation of Spending and is "the same" as one standard deviation of Percentage Taking the Exam.
The full R script follows. Add a comment to this post if this script is useful to you. Enjoy!
# Jags-MultivariateNormal.R
# John Kruschke, November 2015 - June 2017.
# For further info, see:
# Kruschke, J. K. (2015). Doing Bayesian Data Analysis, Second Edition:
# A Tutorial with R, JAGS, and Stan. Academic Press / Elsevier.
# Optional generic preliminaries:
graphics.off() # This closes all of R's graphics windows.
rm(list=ls()) # Careful! This clears all of R's memory!
#--------------------------------------------------------------------------
# Load the data:
#--------------------------------------------------------------------------
myData = read.csv("Guber1999data.csv") # must have file in curr. work. dir.
# y must have named columns, with no missing values!
y = myData[,c("SATT","Spend","PrcntTake")]
#----------------------------------------------------------------------------
# The rest can remain unchanged, except for the specification of difference of
# correlations at the very end.
#----------------------------------------------------------------------------
# Load some functions used below:
source("DBDA2E-utilities.R") # Must be in R's current working directory.
# Install the ellipse package if not already:
want = c("ellipse")
have = want %in% rownames(installed.packages())
if ( any(!have) ) { install.packages( want[!have] ) }
# Standardize the data:
sdOrig = apply(y,2,sd)
meanOrig = apply(y,2,mean)
zy = apply(y,2,function(yVec){(yVec-mean(yVec))/sd(yVec)})
# Assemble data for sending to JAGS:
dataList = list(
zy = zy ,
Ntotal = nrow(zy) ,
Nvar = ncol(zy) ,
# Include original data info for transforming to original scale:
sdOrig = sdOrig ,
meanOrig = meanOrig ,
# For wishart (dwish) prior on inverse covariance matrix:
zRscal = ncol(zy) , # for dwish prior
zRmat = diag(x=1,nrow=ncol(zy)) # Rmat = diag(apply(y,2,var))
)
# Define the model:
modelString = "
model {
for ( i in 1:Ntotal ) {
zy[i,1:Nvar] ~ dmnorm( zMu[1:Nvar] , zInvCovMat[1:Nvar,1:Nvar] )
}
for ( varIdx in 1:Nvar ) { zMu[varIdx] ~ dnorm( 0 , 1/2^2 ) }
zInvCovMat ~ dwish( zRmat[1:Nvar,1:Nvar] , zRscal )
# Convert invCovMat to sd and correlation:
zCovMat <- inverse( zInvCovMat )
for ( varIdx in 1:Nvar ) { zSigma[varIdx] <- sqrt(zCovMat[varIdx,varIdx]) }
for ( varIdx1 in 1:Nvar ) { for ( varIdx2 in 1:Nvar ) {
zRho[varIdx1,varIdx2] <- ( zCovMat[varIdx1,varIdx2]
/ (zSigma[varIdx1]*zSigma[varIdx2]) )
} }
# Convert to original scale:
for ( varIdx in 1:Nvar ) {
sigma[varIdx] <- zSigma[varIdx] * sdOrig[varIdx]
mu[varIdx] <- zMu[varIdx] * sdOrig[varIdx] + meanOrig[varIdx]
}
for ( varIdx1 in 1:Nvar ) { for ( varIdx2 in 1:Nvar ) {
rho[varIdx1,varIdx2] <- zRho[varIdx1,varIdx2]
} }
}
" # close quote for modelString
writeLines( modelString , con="Jags-MultivariateNormal-model.txt" )
# Run the chains:
nChain = 3
nAdapt = 500
nBurnIn = 500
nThin = 10
nStepToSave = 20000
require(rjags)
jagsModel = jags.model( file="Jags-MultivariateNormal-model.txt" ,
data=dataList , n.chains=nChain , n.adapt=nAdapt )
update( jagsModel , n.iter=nBurnIn )
codaSamples = coda.samples( jagsModel ,
variable.names=c("mu","sigma","rho") ,
n.iter=nStepToSave/nChain*nThin , thin=nThin )
# Convergence diagnostics:
parameterNames = varnames(codaSamples) # get all parameter names
for ( parName in parameterNames ) {
diagMCMC( codaObject=codaSamples , parName=parName )
}
# Examine the posterior distribution:
mcmcMat = as.matrix(codaSamples)
chainLength = nrow(mcmcMat)
Nvar = ncol(y)
# Create subsequence of steps through chain for plotting:
stepVec = floor(seq(1,chainLength,length=20))
# Make plots of posterior distribution:
# Preparation -- define useful functions:
library(ellipse)
expandRange = function( x , exMult=0.2 ) {
lowVal = min(x)
highVal = max(x)
wid = max(x)-min(x)
return( c( lowVal - exMult*wid , highVal + exMult*wid ) )
}
for ( varIdx in 1:Nvar ) {
openGraph(width=7,height=3.5)
par( mar=c(3.5,3,2,1) , mgp=c(2.0,0.7,0) )
layout(matrix(1:2,nrow=1))
# Marginal posterior on means:
plotPost( mcmcMat[ , paste0("mu[",varIdx,"]") ] ,
xlab=paste0("mu[",varIdx,"]") ,
main=paste( "Mean of" , colnames(y)[varIdx] ) )
# Marginal posterior on standard deviations:
plotPost( mcmcMat[ , paste0("sigma[",varIdx,"]") ] ,
xlab=paste0("sigma[",varIdx,"]") ,
main=paste( "SD of" , colnames(y)[varIdx] ) )
}
for ( varIdx1 in 1:(Nvar-1) ) {
for ( varIdx2 in (varIdx1+1):Nvar ) {
openGraph(width=7,height=3.5)
par( mar=c(3.5,3,2,1) , mgp=c(2.0,0.7,0) )
layout(matrix(1:2,nrow=1))
# Marginal posterior on correlation coefficient
plotPost( mcmcMat[ , paste0("rho[",varIdx1,",",varIdx2,"]") ] ,
xlab=paste0("rho[",varIdx1,",",varIdx2,"]") ,
main=paste( "Corr. of" , colnames(y)[varIdx1] ,
"and" , colnames(y)[varIdx2] ) )
# Data with posterior ellipse
ellipseLevel = 0.90
plot( y[,c(varIdx1,varIdx2)] , # pch=19 ,
xlim=expandRange(y[,varIdx1]) , ylim=expandRange(y[,varIdx2]) ,
xlab=colnames(y)[varIdx1] , ylab=colnames(y)[varIdx2] ,
main=bquote("Data with posterior "*.(ellipseLevel)*" level contour") )
# Posterior ellipses:
for ( stepIdx in stepVec ) {
points( ellipse( mcmcMat[ stepIdx ,
paste0("rho[",varIdx1,",",varIdx2,"]") ] ,
scale=mcmcMat[ stepIdx ,
c( paste0("sigma[",varIdx1,"]") ,
paste0("sigma[",varIdx2,"]") ) ] ,
centre=mcmcMat[ stepIdx ,
c( paste0("mu[",varIdx1,"]") ,
paste0("mu[",varIdx2,"]") ) ] ,
level=ellipseLevel ) ,
type="l" , col="skyblue" , lwd=1 )
}
# replot data:
points( y[,c(varIdx1,varIdx2)] )
}
}
# Show data descriptives on console:
cor( y )
apply(y,2,mean)
apply(y,2,sd)
#-----------------------------------------------------------------------------
# Difference of correlations. Change the specification of indices as
# appropriate to your variable names and interests.
#-----------------------------------------------------------------------------
# Specify indices of desired correlations:
rhoAidx1 = which(colnames(y)=="SATT")
rhoAidx2 = which(colnames(y)=="Spend")
rhoBidx1 = which(colnames(y)=="SATT")
rhoBidx2 = which(colnames(y)=="PrcntTake")
# Make plot of posterior difference:
openGraph(height=3.5,width=4)
par( mar=c(3.5,1,2,1) , mgp=c(2.0,0.7,0) )
plotPost( mcmcMat[ , paste0("rho[",rhoAidx1,",",rhoAidx2,"]") ]
- mcmcMat[ , paste0("rho[",rhoBidx1,",",rhoBidx2,"]") ],
xlab=paste0(
"rho[",rhoAidx1,",",rhoAidx2,"] - rho[",rhoBidx1,",",rhoBidx2,"]" ) ,
main=bquote( rho[ list( .(colnames(y)[rhoAidx1]) ,
.(colnames(y)[rhoAidx2]) ) ]
- rho[ list( .(colnames(y)[rhoBidx1]) ,
.(colnames(y)[rhoBidx2]) ) ] ) ,
cex.main=1.5 , ROPE=c(-0.1,0.1) )
#---------------------------------------------------------------------------